We have practical experience in analysing data from different sequencing techniques (short reads, PacBio, Oxford nanopore), and we offer customised data analysis and visualization to help you make the most out of your genomic data.
What kind of analysis do we do
Methods:
- Genome assembly
- SNP calling & GWAS
- Hi-C, Micro-C, 4C-seq
- (sc)ATAC-seq & multiomics
- ChIP-seq
- DNA methylation
- Metagenomic
- DNA-FISH
- NeoAg prediction
- CRISPR-Cas9
Some examples:
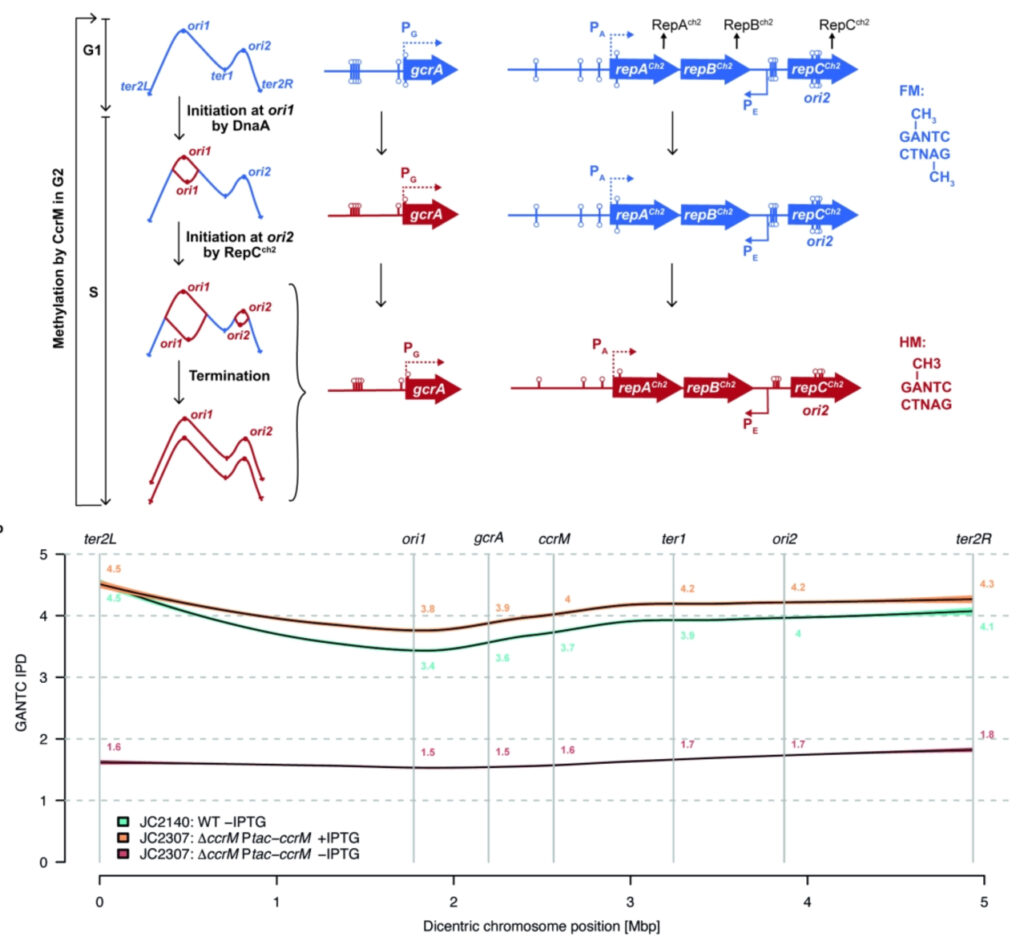
DNA methylation by CcrM contributes to genome maintenance in the Agrobacterium tumefaciens plant pathogen
In this study, we analysed the CcrM-dependent methylation of GANTC motifs on the A. tumefaciens dicentric chromosome, showing that the vast majority of the 5166 double-stranded GANTC motifs found on the dicentric chromosome of JC2307 cells are in a non-methylated state after 7 hours of CcrM depletion (red line), but that it is restored to levels comparable to wild type upon re-induction of CcrM. We could also show that replication origin ori2 initiates with a significant delay during the A. tumefaciens cell cycle when its dicentric chromosome is hypo-methylated due to a depletion of CcrM
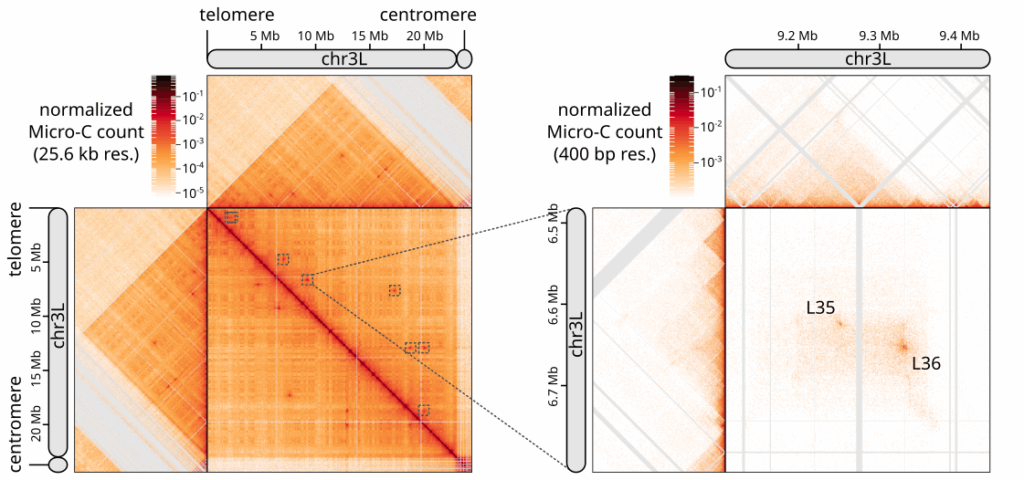
Chromosome-level organization of the regulatory genome in the Drosophila nervous system
In this publication, we prepared Hi-C and Micro-C plots and developed a custom algorithm for meta-loop identification in larval CNS contact maps.
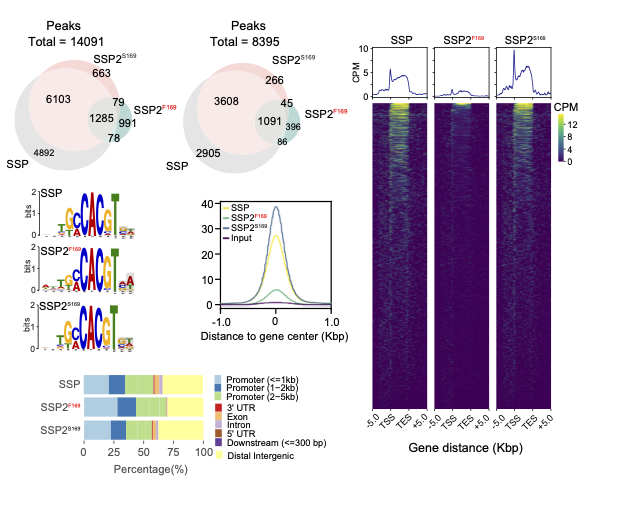
Repairing a deleterious domestication variant in a floral regulator of tomato by base editing
In this study, we helped investigate the load of deleterious mutations in various tomato cultivars by analysing genomic variants, DAP-seq, RNA-seq and CRISPR-Cas9, demonstrating that deleterious mutations in the bZIP transcription factor SSP2 (a paralog of SSP, both well evolutionary conserved) affected the inflorescence.
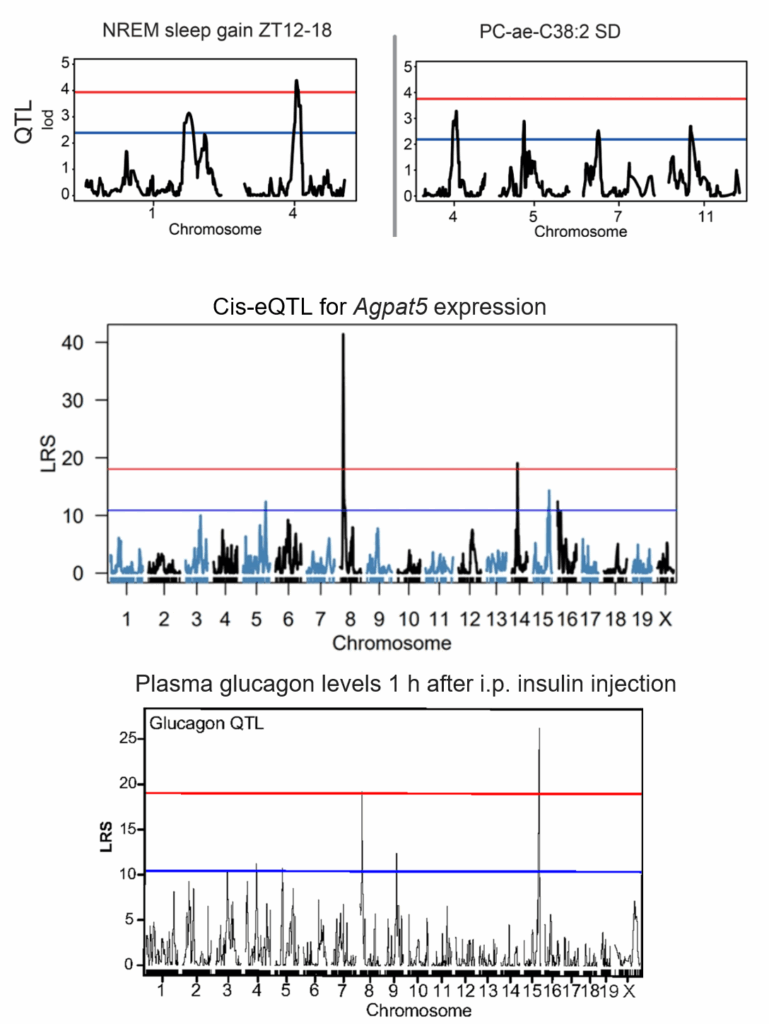
Quantitative trait locus (QTL) mapping in the BXD mouse panel
In these publications, we performed quantitative trait locus mapping the in the BXD mouse recombinant panel.
Examples of the phenotypes analyzed:
NREM sleep gain and phosphatidylcholine level.