What kind of analysis do we do
Methods:
- Mathematical Modelling
- Signal annotation using Machine learning
- Electropherogram
- EEG spectra analysis
Some Examples:
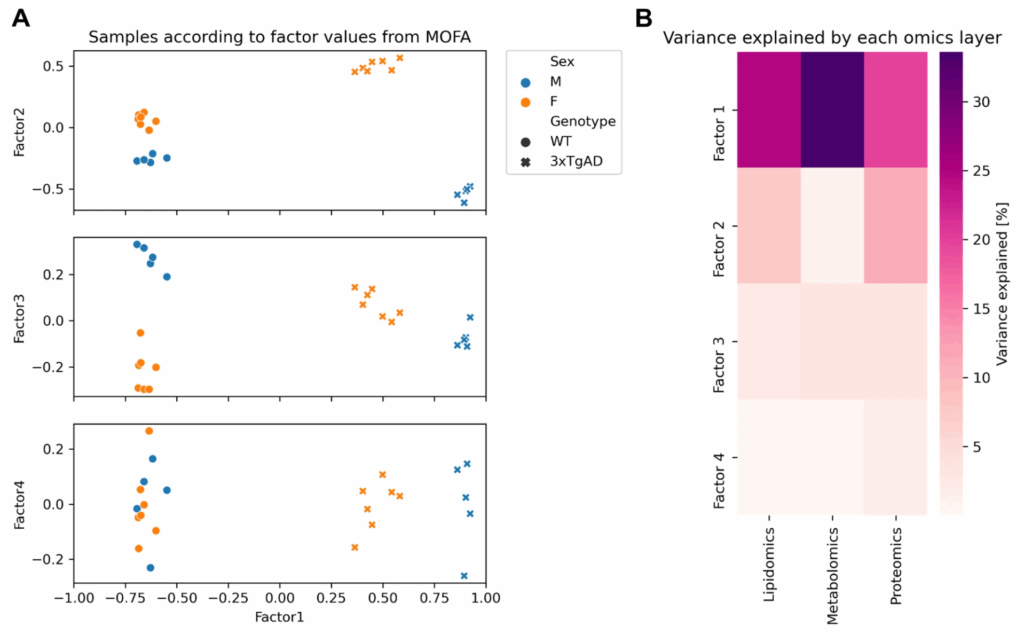
Molecular insights into sex-specific metabolic alterations in Alzheimer’s mouse brain using multi-omics approach
In this publication, we performed multi-omics factor analysis (MOFA) to identify the shared factors between lipidomics, metabolomics and proteomics datasets in an Alzheimer mouse model. Strong differences were found between male and female mice that carried 3xTg-AD (three mutant alleles; homozygous for the Psen1 mutation and homozygous for the co-injected APPSwe and tauP301L transgenes)
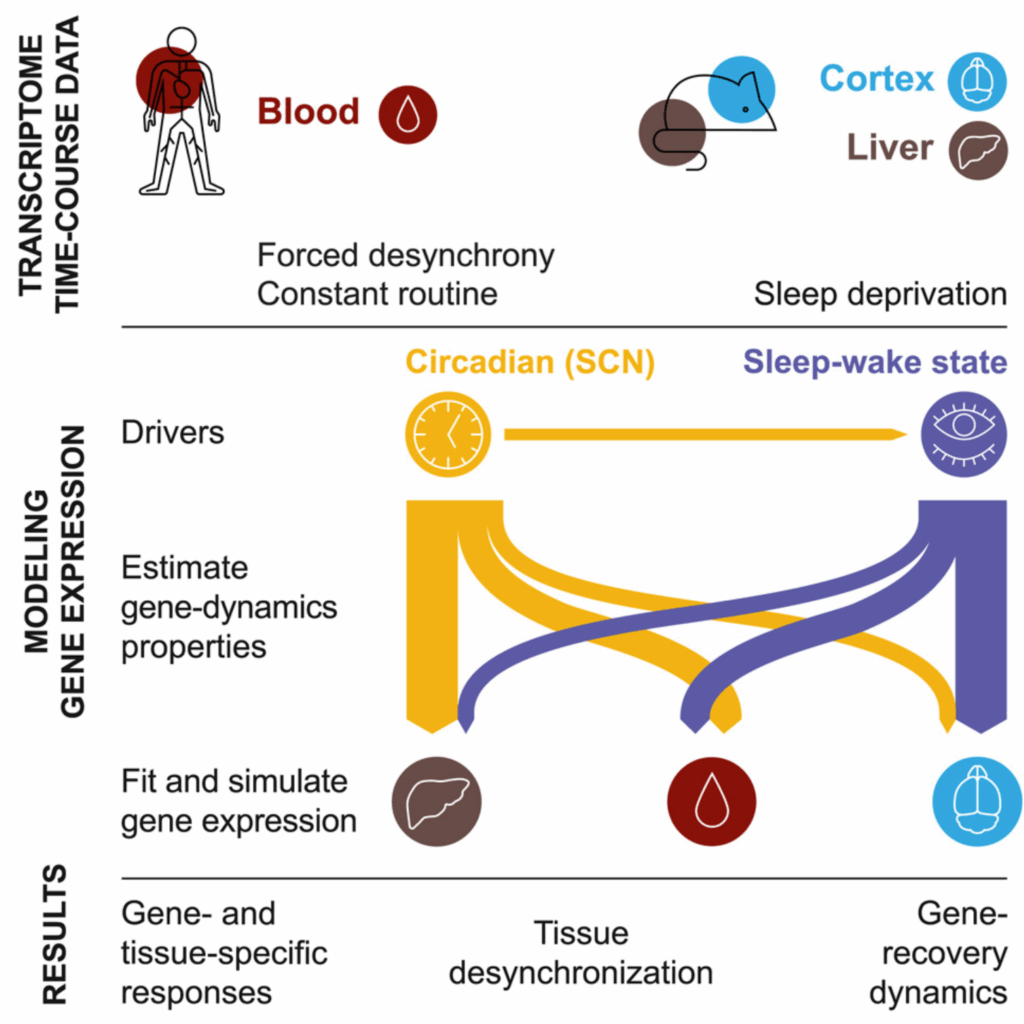
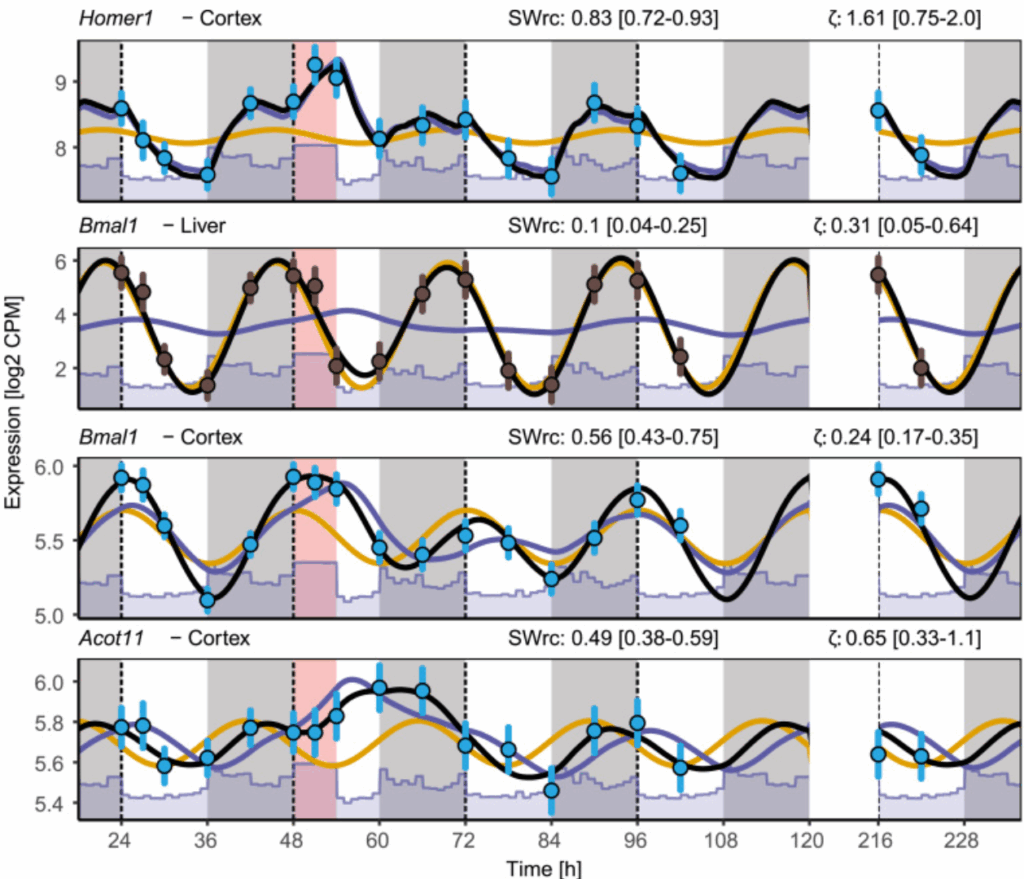
Model integration of circadian- and sleep-wake-driven contributions to rhythmic gene expression
Daily rhythmic gene expression can be driven by multiple factors. In this publication, we disentangle the contribution of circadian rhythms and sleep-wake history to the rhythms of mouse cortical and liver transcriptomes as well as human transcriptome.
Below, the rhythm of 4 genes in mice after a sleep deprivation (red box), with corresponding fit from our model (black line). The model is composed of a sleep-wake (purple) and circadian (orange) contribution. The sleep-wake contribution (SWrc) can be different for the same genes in different tissue (e.g. Bmal1), while damping ratio (ζ), a parameter of the model, is similar.
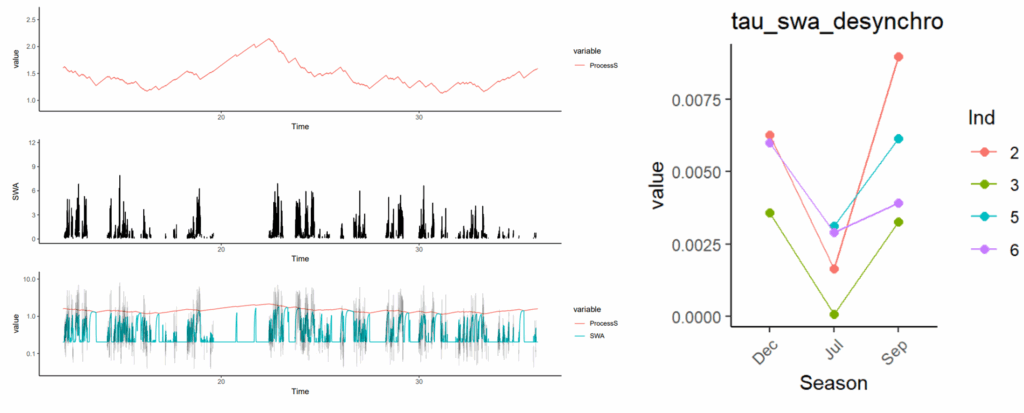
Model slow waves activity (SWA) and Process-S in arctic reindeers
In this publication, we helped modeling sleep pressure (process-S) during NREM sleep and rumination in arctic reindeer. We modeled that rumination help reducing sleep pressure and identify differences in model parameters depending on seasons.
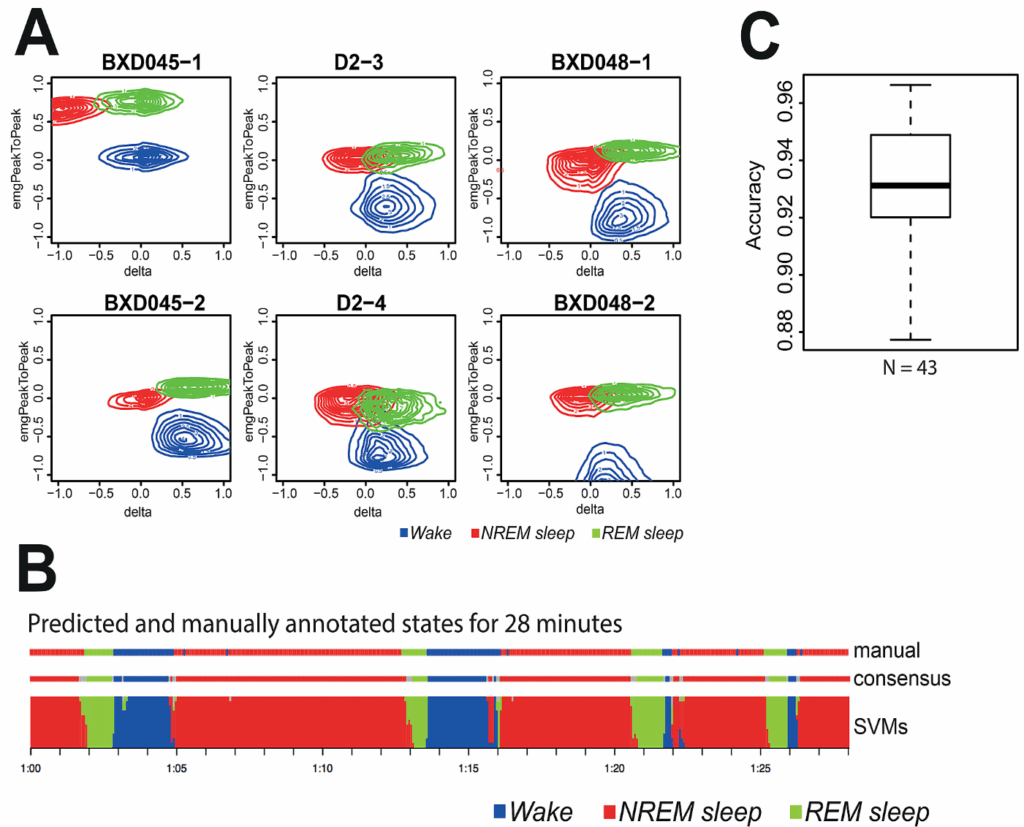
Supervised machine learning approach to score sleep states in recombinant inbred mice panel
In the course of the project, a total of 260 mice electroencephalograms (EEG) and electromyogram (EMG) have been recorded during 4 days to evaluate differences in quality of sleep between mouse strains. The usual analysis procedure entails a visual scoring of each 4s epoch to determine the behavioral state of the mouse (Wake, NREM, or REM). However, in our case, this time-consuming process was not applicable. Thus each EEG and EMG spectra were processed using a Fast Fourier Transform (FFT), to export a total of 203 numerical parameters. Various combinations of these parameters were used to train a series of support vector machines (SVM) specifically tuned for each mouse to reduce the time necessary to perform visual scoring.