Fine analysis of various protein structures, either experimentally obtained or through protein modelling can inform about the potential impact of various rare variants and mode of action, as showcased in the few selected examples below.
What kind of analysis do we do
Methods:
- 3D models
- Protein interactions analysis
- Mutations impact
- Low homology modelling
- Structure comparison
Some examples:
MCTS2 and distinct eIF2D roles in uORF-dependent translation regulation revealed by in vitro re-initiation assays
MCTS2 is not a pseudogene, and residues that differ from MCTS1 (in cyan on the image below) are all located on the same side of the protein and solvent accessible (in red on the image below).
Hover on the image to see how EIF2D (in teal) can occupy the same space as the DENR-MCTS2 complex.
Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles
Variants that affect residues of the DEGRON domain will prevent proper degradation of AFF3, which results in a deleterious accumulation of the protein leading to abnormal development.
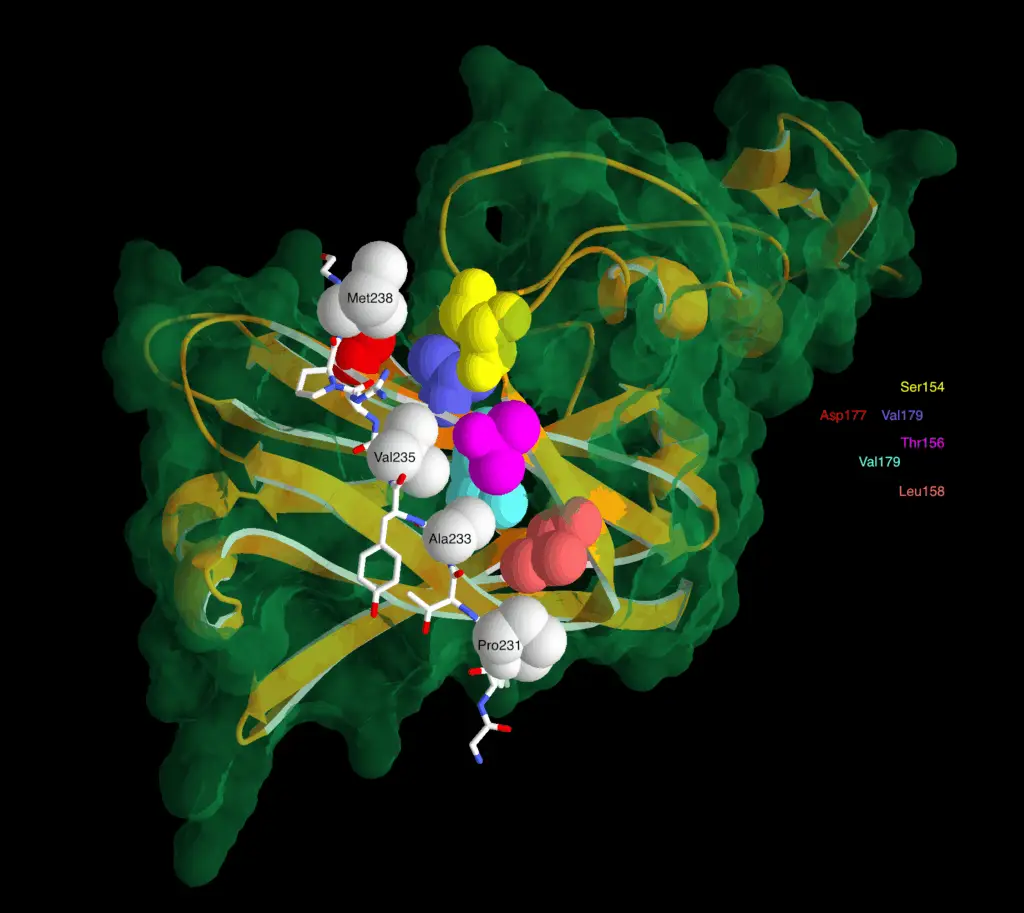
Mutation-specific pathophysiological mechanisms define different neurodevelopmental disorders associated with SATB1 dysfunction
Mutations in SATB1 can lead to neurodevelopmental disorders. In particular, we report variants located at structurally equivalent positions in the CUT1 and CUT2 DNA binding domains of SATB1, which affect DNA binding properties.
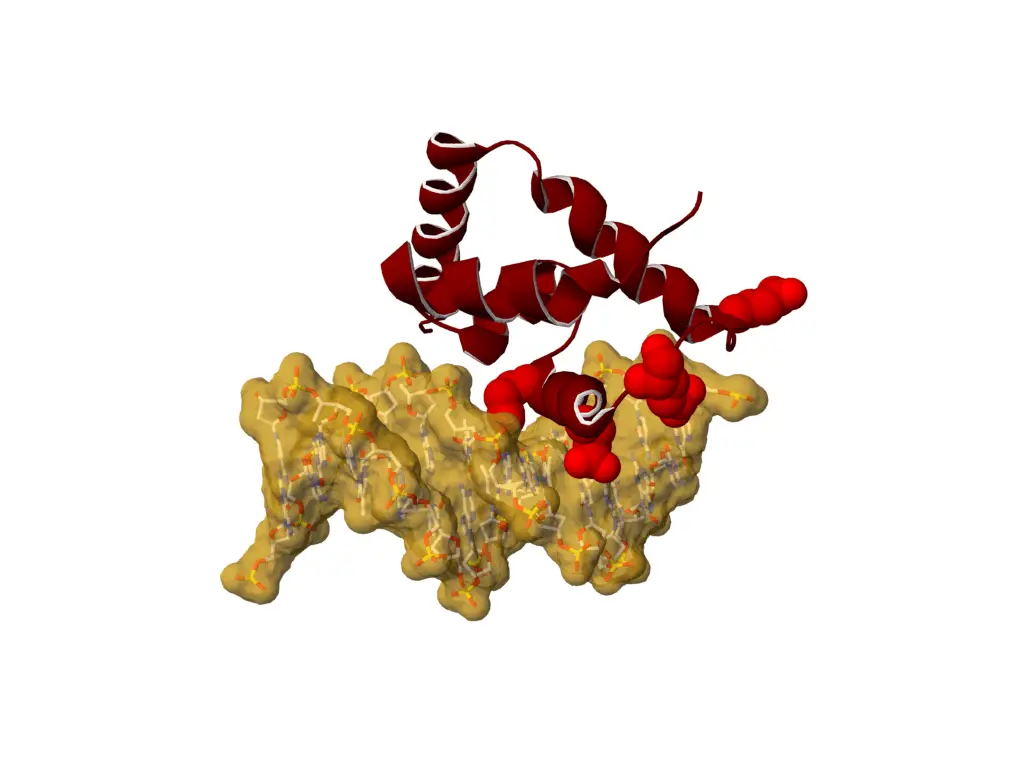
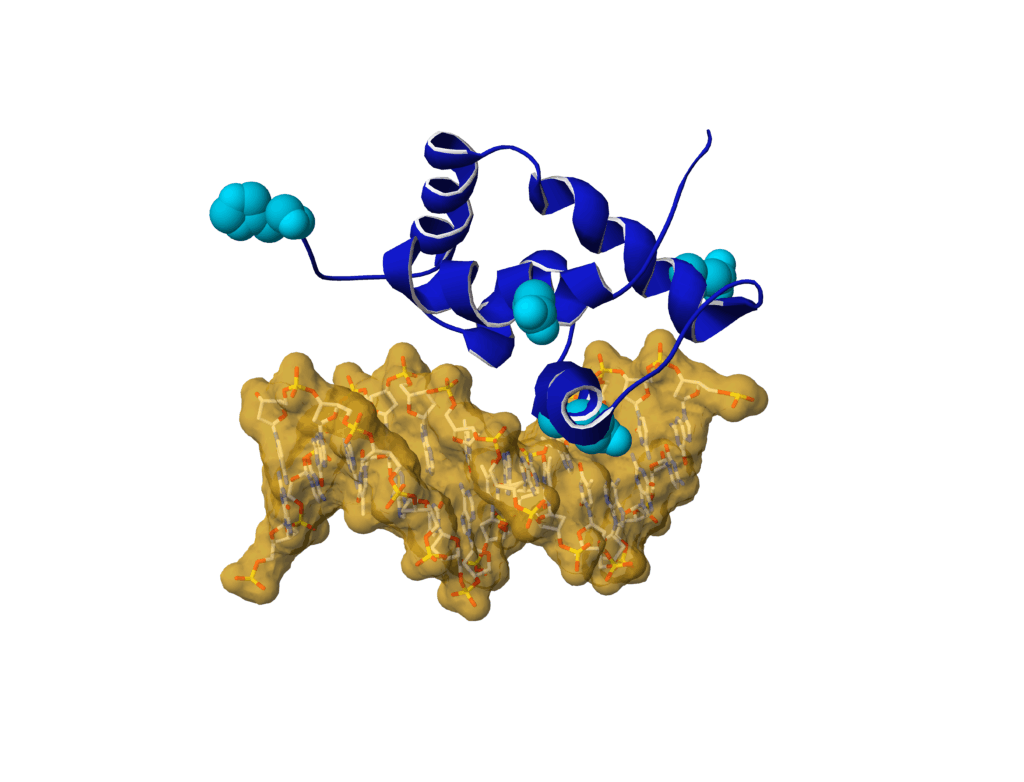
Variants in USP48 encoding ubiquitin hydrolase are associated with autosomal dominant non-syndromic hereditary hearing loss
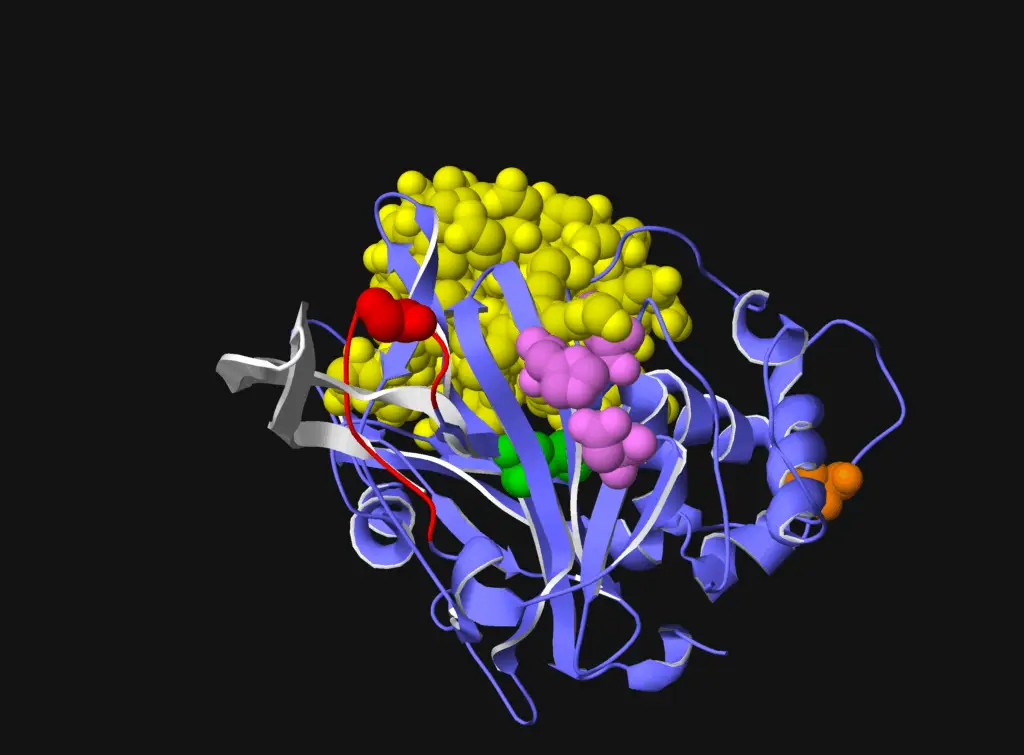
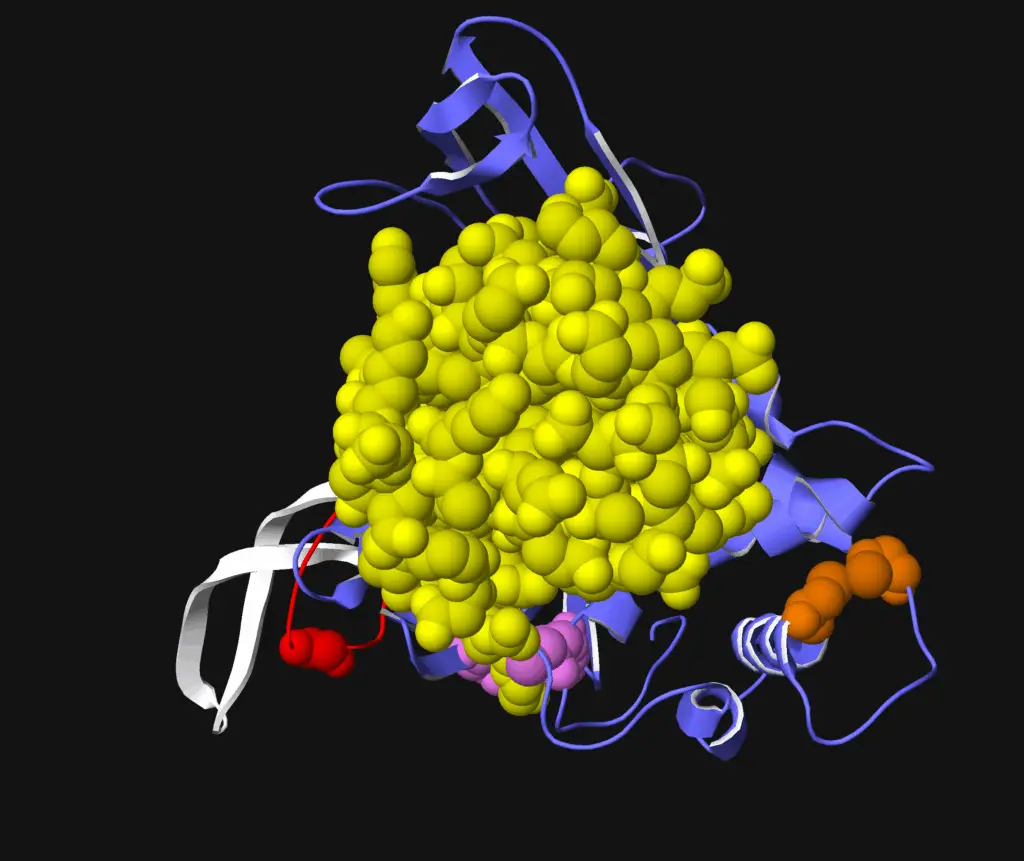
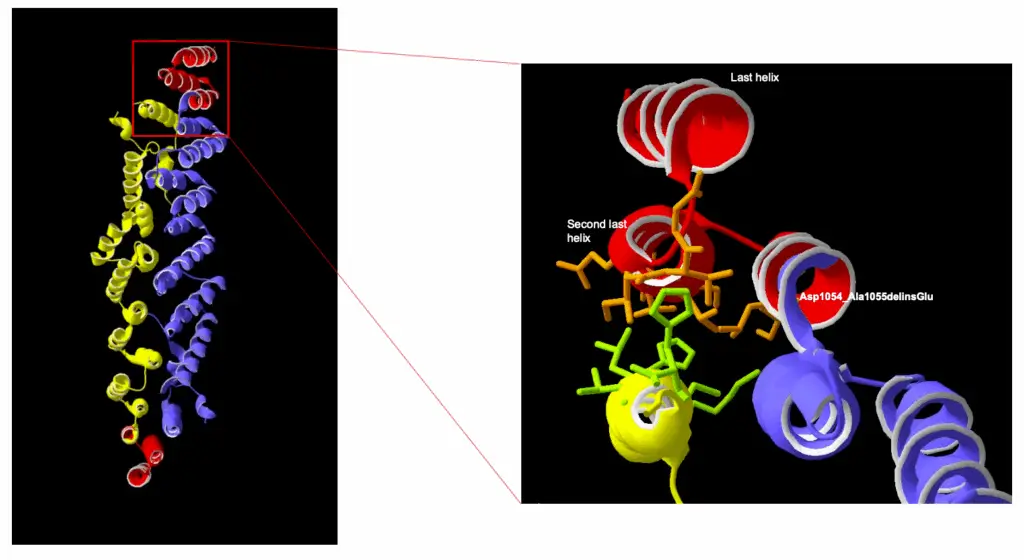
A Biallelic Truncating Variant in the TPR Domain of GEMIN5 Associated with Intellectual Disability and Cerebral Atrophy
GEMIN5 is a multifunctional RNA-binding protein required for the assembly of survival motor neurons. Several bi-allelic truncating and missense variants in this gene are reported to cause a neurodevelopmental disorder characterized by cerebellar atrophy, intellectual disability, and motor dysfunction. In this article, we discuss (Asp1054_Ala1055delinsGlu), a novel GEMIN5 pathogenic variant
Software
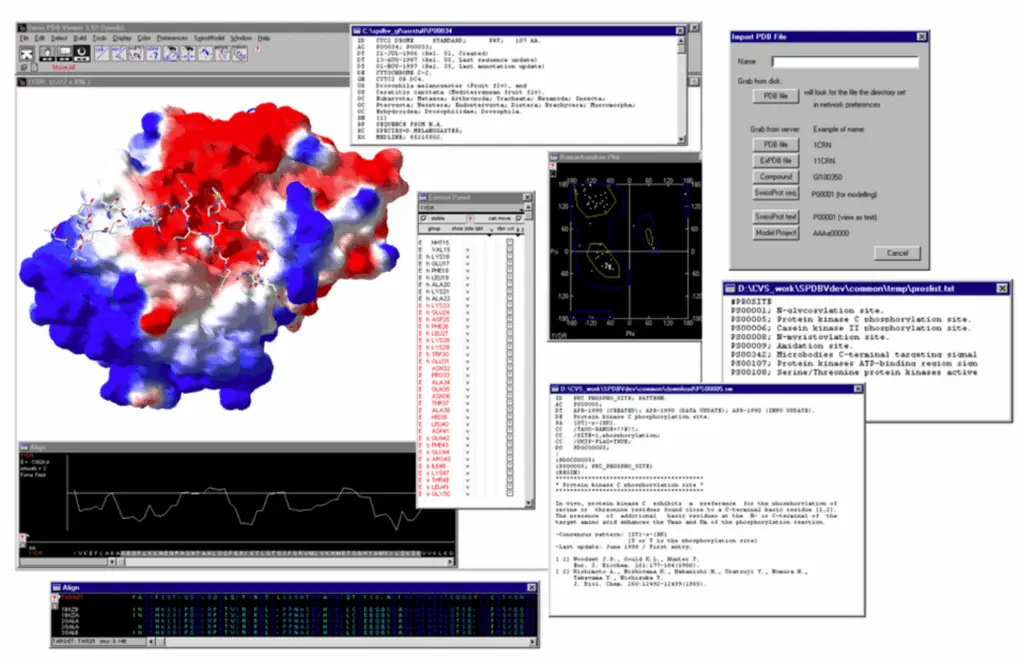
All the examples above have been analysed with Swiss-PdbViewer (aka DeepView), an application that provides a user friendly interface allowing to analyze several proteins at the same time. The proteins can be superimposed in order to deduce structural alignments and compare their active sites or any other relevant parts. Amino acid mutations, H-bonds, angles and distances between atoms are easy to obtain thanks to the intuitive graphic and menu interface.
This software, whose development started in 1995, has been used by the author and the community to explore the potential effect of SNVs, perform low homology modelling and propose mutations to test in the laboratory.