What kind of tools do we develop
- explore and visualize datasets to be published
- to facilitate and automate the execution of analysis pipeline
- to provide reproducible solutions through the usage of Docker in the backend
- to make tools normally accessible only at the command line accessible to anyone
All these solutions have in common to be accessible on internet which ensures accessibility and flexibility in the presentation of the results.
Online analysis pipelines
ASAP - Automated Single-cell Analysis Portal
![]()
ASAP is an open, scalable and interactive web-based portal for (single-cell) omics analyses. It allows biologists to do analyses from the count matrix to the collaborative annotation without any coding. It includes all the steps of traditional single-cell dataset analysis, such as cell filtering, gene filtering, normalization, scaling, removal of covariates, dimension reductions, clustering, differential expression analysis and gene enrichments. Users also have the ability to download all the code and to locally reproduce their analyses within a docker environment.
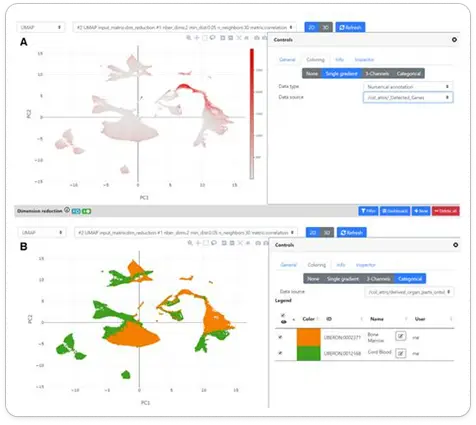
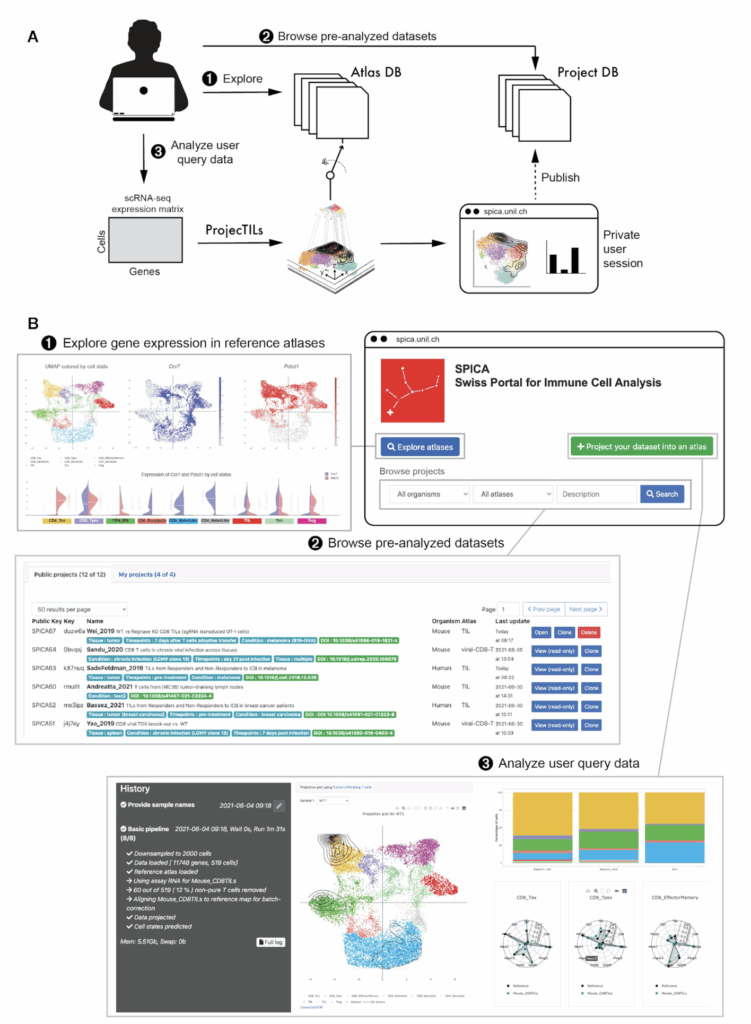
SPICA - Swiss Portal for Immune Cell Analysis
We developed the front end of SPICA, a web resource dedicated to the exploration and analysis of single-cell RNA-seq data of immune cells
Data visualization & exploration
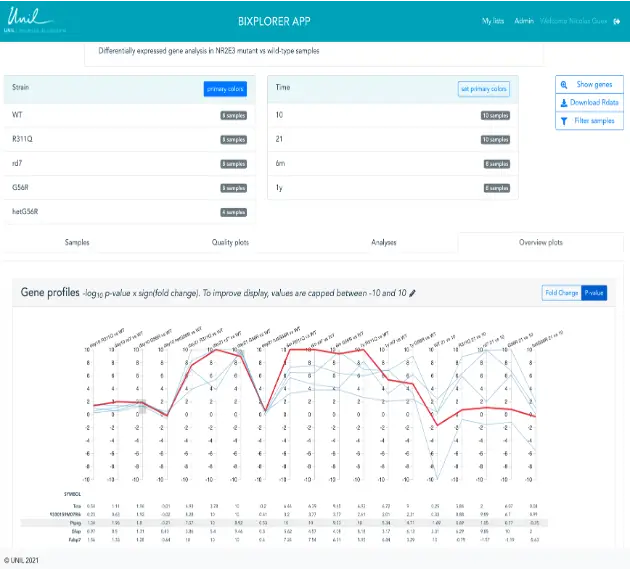
Bixplorer - visualization of omics datasets and analyses
Upon request, we can load RNAseq or proteomics differential expression analyses results into Bixplorer. Our web tool provides a way to mine RNAseq and proteomics results to rapidly check expression profiles of significant genes / proteins, and / or select transcripts / proteins that are significantly up or down-regulated only under specific conditions.
For example, this tool was used to identify “Transcriptomic Signature Differences Between SARS-CoV-2 and Influenza Virus Infected Patients.” Front. Immunol. (2021)
ClockProfile - visualization of the rythmicity of gene expression
ClockProfile is a web-application providing an easy-to-use interface for temporal RNA-Seq datasets. Currently, it allows the user to visualize the temporal gene expression in liver of gene sets and individual genes in several mouse KO models of clock genes (i.e. Bmal1 KO, Cry1/2 dKO and PARbZip KO. Raw RNA-Seq data is accessible at GEO (GSE135898 and GSE135875).

Swiss-BXD - interactive visualization of a systems genetics database
We used a systems genetics approach in the BXD mouse strains to assembled a comprehensive experimental knowledge base comprising a deep “sleep-wake” phenomenon, central and peripheral transcriptomes, and plasma metabolome data, collected under undisturbed baseline conditions and after sleep deprivation (SD). We created an analytical tool website to interactively interrogate the database, visualize the molecular networks altered by sleep loss, and prioritize candidate genes.
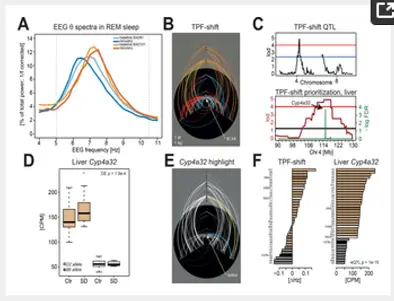
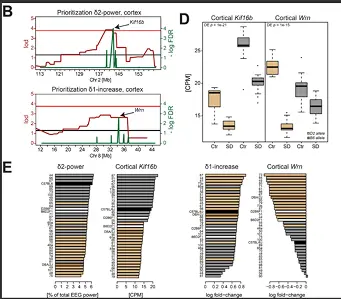
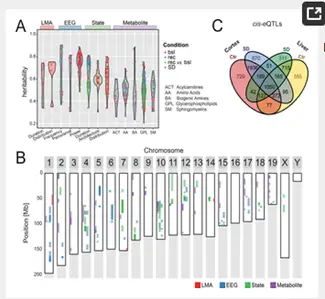
Databases
SwissPalm - database on protein S-palmitoylation

SwissPalm is an open, comprehensive, manually curated database to study protein S-palmitoylation.
Mycobrowser - database on mycobacteria species

Mycobrowser (Mycobacterial browser) is a comprehensive genomic and proteomic data repository for pathogenic mycobacteria. It provides manually-curated annotations and appropriate tools to facilitate genomic and proteomic study of these organisms.
GetPrime - database of primers for qPCR

GETPrime is a primer database supported by a novel platform that uniquely combines and automates several features critical for optimal qPCR primer design.
Customized data management for platforms
PCFpro - LIMS of the Proteomics Core Facility of EPFL
PCFpro is a data management and billing platform tailored specifically for the Proteomics Core Facility of EPFL.
MetaboApp - LIMS of the Metabolomics Unit of UNIL
MetaboApp is a data management and data analysis platform created specifically for the Metabolomics Unit of UNIL.
CloneMap - database of clones available at the Gene Expression Core Facility of EPFL
LIMS of the Lausanne Genomic Technologies Facility (LGTF) of UNIL
The backend API of the existing LIMS was reimplemented from Scala to Python using Django. The migration is completely transparent to end users.
Data storage management
Pangea - long-term storage of files and associated metadata on S3
Pangea is a tool that allows members of a lab to annotate files being part of experiments, and to archive them together with the metadata on a long-term s3 storage.
Hi-TIDe knowledge management system
This application tracks the lab work from 100 users in 9 laboratories.
It also provides a customized integration of results from more than 20 modalities.

